I run an independent research group and teach in the School of Chemical Sciences at the University of Auckland, New Zealand.
My group studies how nature performs chemistry using enzymes, and how we can harness that understanding to solve real-world problems.
See my profile page here: https://profiles.auckland.ac.nz/tristan-de-rond
scroll down to see a list of my publications
My current interests include:
- Genomics of biosynthetically "gifted" marine organisms
- Metabolic pathway and enzyme discovery
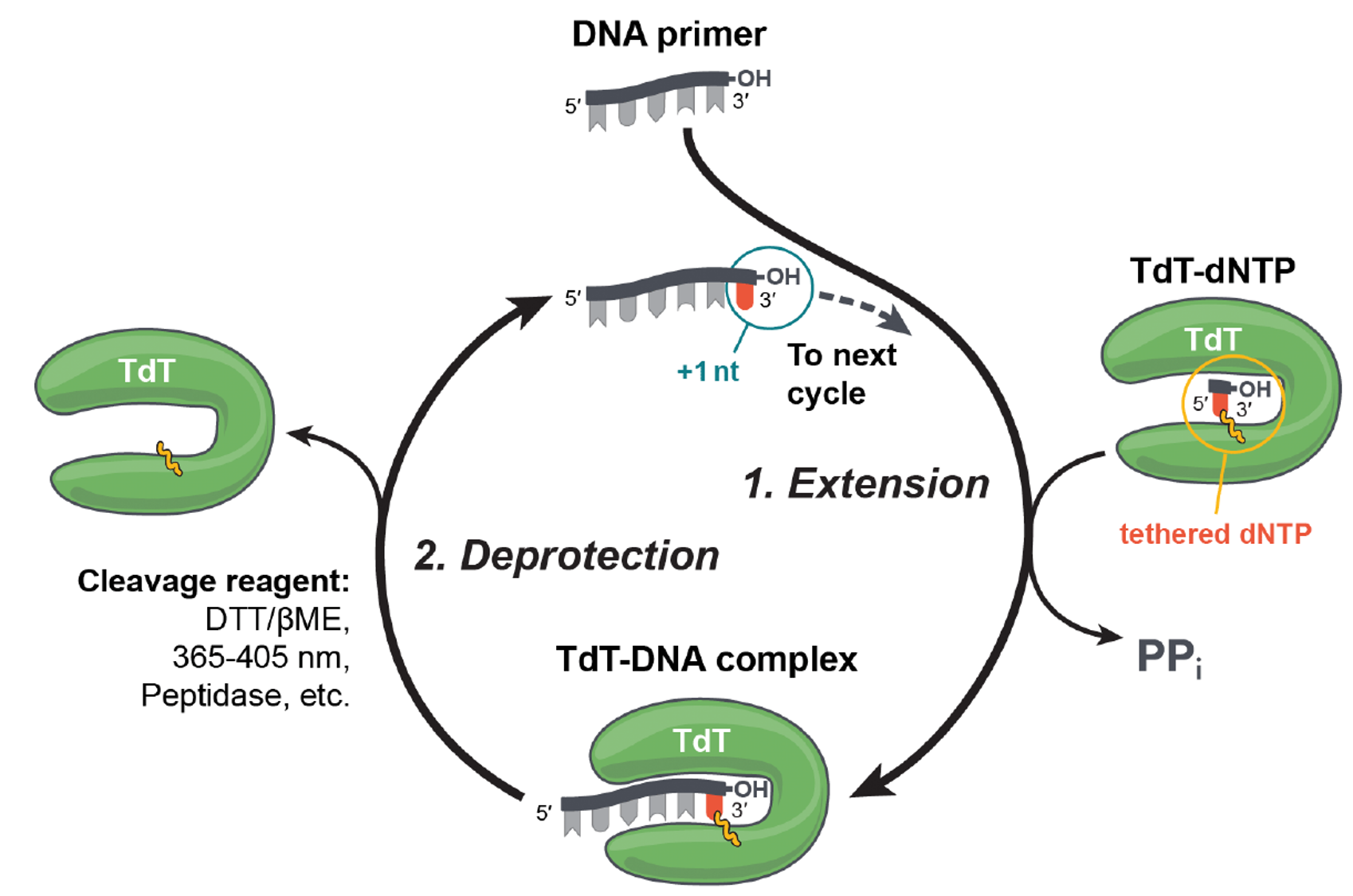
- "Biosynthetic methodology"
- Biocatalysis
- Metabolic engineering
- High-throughput analytical chemistry
- Laboratory automation
- Graduate and undergraduate student mentorship
Tristan's Tips and Tricks
07.09.2020
Chemistry software
05.04.2019
Wet Lab Tips and Tricks
03.04.2019
Uncategorised
Peer-reviewed publications
If you're unable to access a paper you want to read, please contact me and I can share an electronic copy for personal use.

- Wong J, de Rond T, d’Espaux L, et al., (2018). High-titer production of lathyrane diterpenoids from sugar by engineered Saccharomyces cerevisiae. Metab. Eng. 45.
- Curran S., Hagen, A., Poust, S. , Chan L.J.G., Garabedian B.M., de Rond, T. et al. (2018) Probing the Flexibility of an Iterative Modular Polyketide Synthase with Non-Native Substrates in Vitro. ACS Chem Biol.
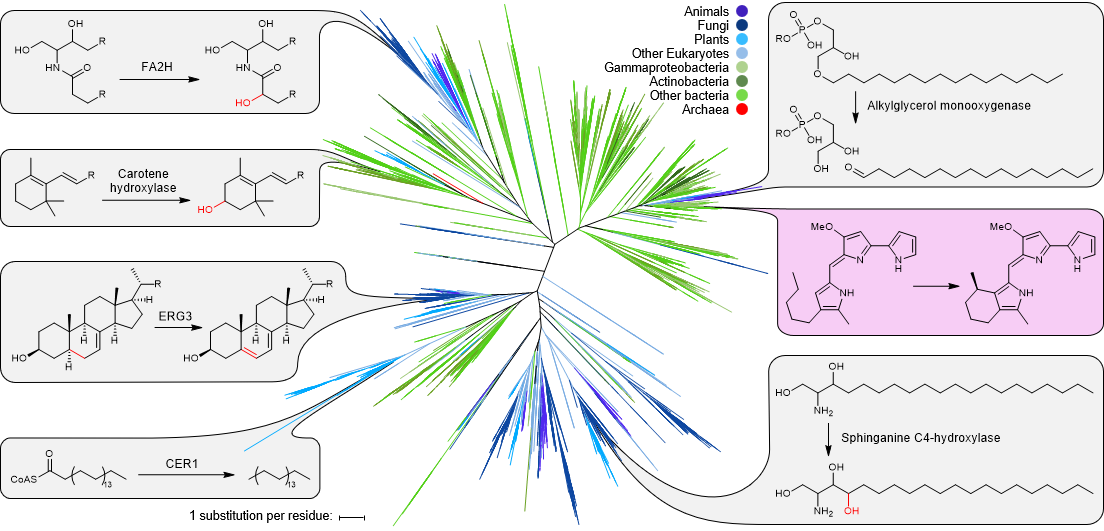
- de Rond, T., Stow, P., Eigl, I., et al. (2017) Oxidative cyclization of prodigiosin by an alkylglycerol monooxygenase-like enzyme. Nat. Chem. Biol. 13
-
de Raad, M.*, de Rond, T.*, Rübel, O., et al. (2017). OpenMSI Arrayed Analysis Toolkit: Analyzing spatially defined samples using mass spectrometry imaging. Anal. Chem. 89 (code available at https://github.com/biorack/omaat)
-
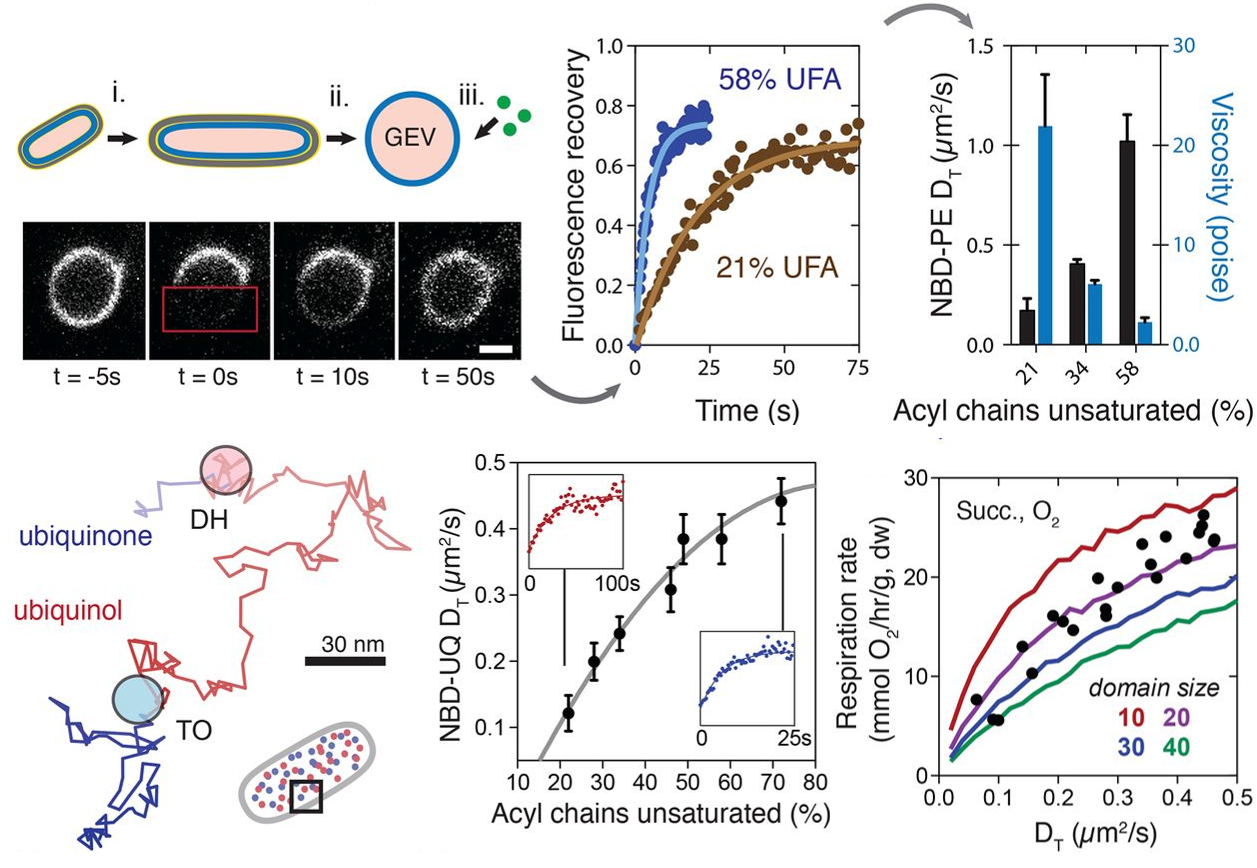
Degreif, D., de Rond, T., Bertl, A. Keasling, J.D., and Budin, I. (2017). Lipid engineering reveals regulatory roles for membrane fluidity in yeast flocculation and oxygen-limited growth. Metab. Eng. 41
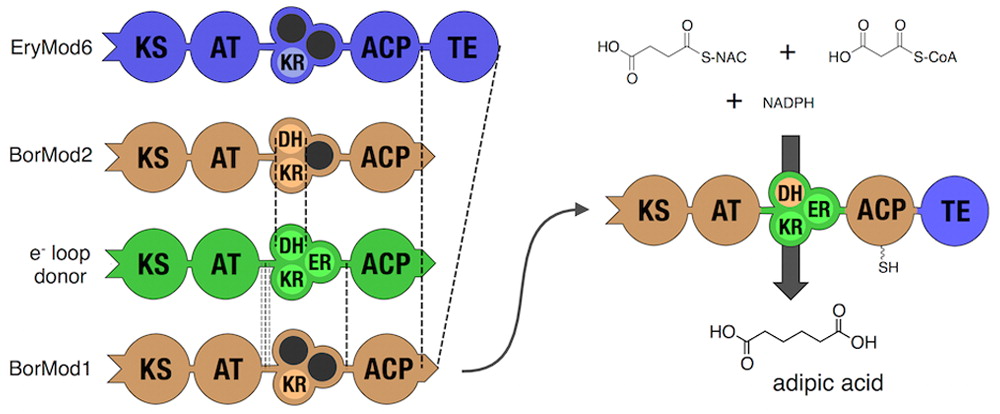
- Hagen, A., Poust, S., de Rond, T., Fortman, J.L., Katz, L., Petzold, C.J., and Keasling, J.D. (2015). Engineering a polyketide synthase for in vitro production of adipic acid. ACS Synth. Biol. 2
- Johnson, R.E., de Rond, T., Lindsay, V.N.G., Keasling, J.D., and Sarpong, R. (2015). Synthesis of Cycloprodigiosin Identifies the Natural Isolate as a Scalemic Mixture. Org. Lett. 17
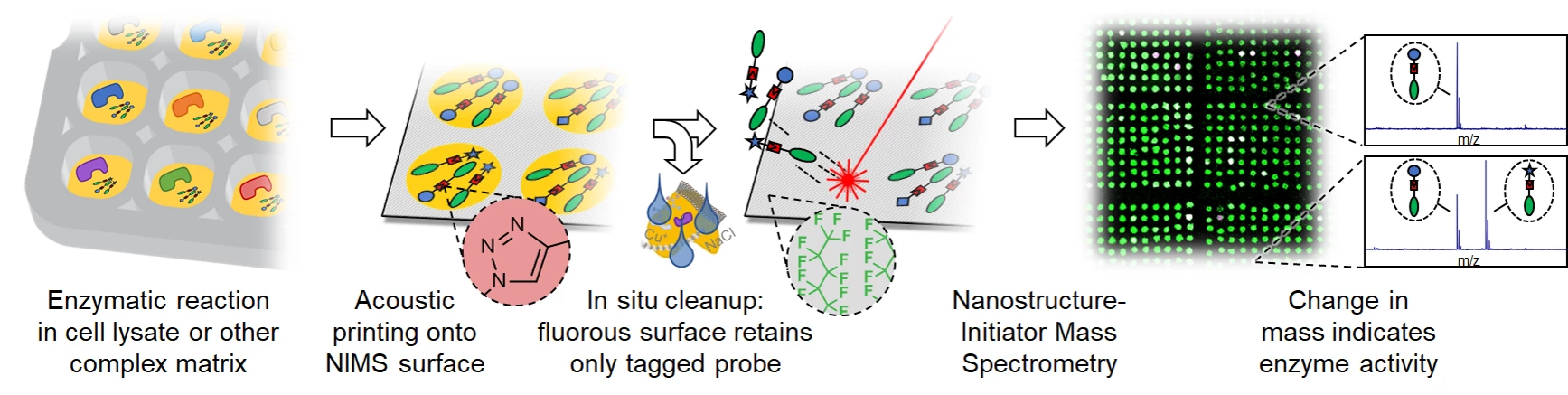
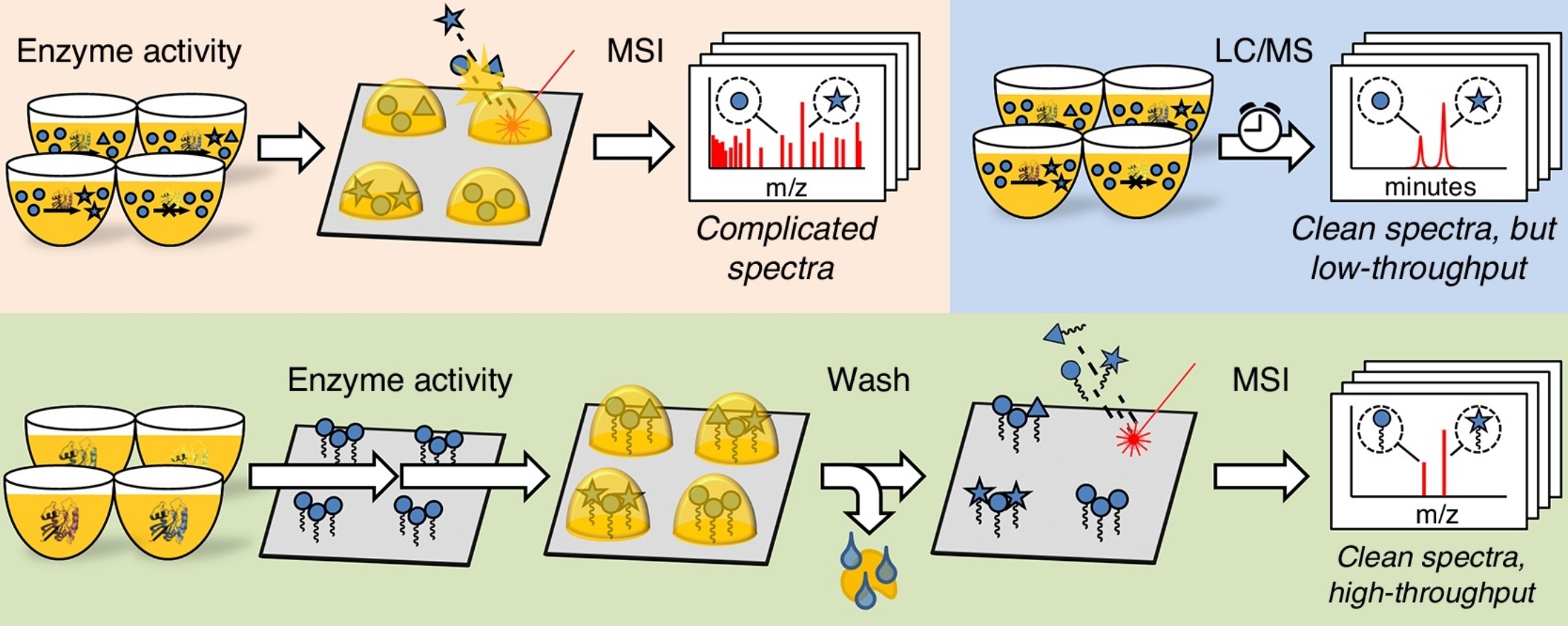
- de Rond, T., Danielewicz, M., and Northen, T. (2015). High throughput screening of enzyme activity with mass spectrometry imaging. Curr. Opin. Biotechnol. 31
- Hagen, A., Poust, S., de Rond, T., Yuzawa, S., Katz, L., Adams, P.D., Petzold, C.J., and Keasling, J.D. (2014). In vitro analysis of carboxyacyl substrate tolerance in the loading and first extension modules of borrelidin polyketide synthase. Biochemistry 53
- de Rond, T., Peralta-Yahya, P., Cheng, X., Northen, T.R., and Keasling, J.D. (2013). Versatile synthesis of probes for high-throughput enzyme activity screening. Anal. Bioanal. Chem. 405